A simple “scatterplot”
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_point()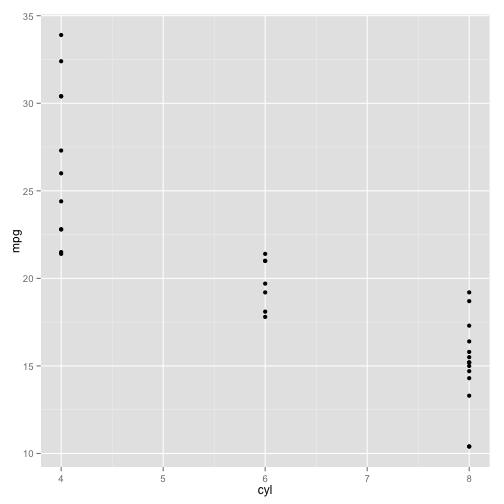
Kodi Weatherholtz
kweatherholtz@ling.ohio-state.edu
“ggplot2 is a plotting system for R, based on the grammar of graphics, which tries to take the good parts of base and lattice graphics and none of the bad parts. It takes care of many of the fiddly details that make plotting a hassle (like drawing legends) as well as providing a powerful model of graphics that makes it easy to produce complex multi-layered graphics.”
Hadley Wickham, developer, http://ggplot2.org/
“ggplot2 is designed to work in a layered fashion, starting with a layer showing the raw data then adding layers of annotations and statistical summaries.”
Wickham, 2009, ggplot2: Elegant Graphics for Data Analysis
in base R:
– graphics are composed of simple raw elements (e.g., points, lines)
– it is difficult to add new components to existing graphics (e.g., error bars, legends)
ggplot:
– graphics involve high-level elements (representations of raw data, statistical transformations)
– elements are easily combined to form complex graphics
## even if you already have ggplot installed, you should reinstall it, as some
## function and argument names have changed with the release of version 9.2
# install.packages('ggplot2')
require(ggplot2)Basic Examples
a data frame
a set of aesthetic mappings
– describe how variables in a data frame are mapped to graphical attributes
– x- and y-axis variables, colo(u)rs, subset groupings, linetypes
one or more geometric objects , called geoms
– determine how values are rendered graphically
– points, lines, boxplots, bars, etc.
statistical transformations, called stats
– e.g., regression lines, bootstrapped confidence intervals, log transform of scale and coordinate systems
one or more facets to create lattice/trellis plots
– e.g., plot each subject’s data individually
a coordinate system, or coord
– default is Cartesian coordinates, but other systems can be specified
– e.g., polar coords, log transformed coords, map projections (latitude and longitude)
scales
– e.g., adjust size of points in a scatterplot to be proportional on a specified scale
# mtcars is a built in data set.
head(mtcars) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
# Let's tidy it up a bit
# turn rownames (car model) into vector, and select a subset of columns
df1 <- data.frame(model = as.factor(row.names(mtcars)),
mtcars[,c(11,1:2,4,6)])
row.names(df1) <- NULL
head(df1) model carb mpg cyl hp wt
1 Mazda RX4 4 21.0 6 110 2.620
2 Mazda RX4 Wag 4 21.0 6 110 2.875
3 Datsun 710 1 22.8 4 93 2.320
4 Hornet 4 Drive 1 21.4 6 110 3.215
5 Hornet Sportabout 2 18.7 8 175 3.440
6 Valiant 1 18.1 6 105 3.460ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_point()
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_boxplot()
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin()
# adjust formatting of x- and y- axes.
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin() +
# change font size, font color, and axis label position
theme(axis.text = element_text(colour="black", size=15),
axis.title.x= element_text(face="bold", size=18, vjust=-0.45),
axis.title.y = element_text(size=18, vjust=.2),
axis.ticks = element_line(colour="red"))
my.theme <- theme(axis.text = element_text(colour="black", size=15),
text = element_text(size=16),
title = element_text(size=19),
axis.title.x= element_text(vjust=-0.45),
axis.title.y = element_text(vjust=.2),
axis.ticks = element_line(colour="black"),
axis.line = element_line())ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin() +
geom_point() + # add a layer showing raw data points
my.theme # use the custom theme we just defined
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin() +
geom_point(position=position_jitter(width=.1)) + # add jitter to points
my.theme
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin() +
geom_dotplot(binaxis = "y", stackdir = "center") + # specify which axis to use for 'binning'
my.theme
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_violin() +
geom_dotplot(binaxis = "y", stackdir = "center", dotsize = .65) + # change size of dots
my.theme
Basic Embellishments
by definition, boxplots display median and interquartile ranges, but not means
– ggplots are built in layers, so it’s easy to add a layer for descriptive statistics like means
scatterplots display values for multiple continuous variables
– it’s also easy to add a regression line to show direction and size of correlation
all plots can be annotated with text
ggplot(data = df1, aes(x = cyl, y = mpg, group = cyl)) +
geom_boxplot() +
# fun.y=mean calculates the mean of the y-axis variable (mpg) for each group.
# geom="point" indicates means should be plotted as points over top of the boxplots.
stat_summary(fun.y = mean, geom = "point", color = "red", size = 3) +
my.theme
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() + # make scatterpot
geom_smooth() + # add loess line
scale_x_continuous("gross horsepower") + # change axis labels and limits
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
my.theme
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
geom_smooth(se = FALSE) + # remove error ribbon
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
my.theme
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
stat_smooth(geom = "pointrange") + # note the change to stat_smooth()
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
my.theme
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y~(x)) +
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
my.theme
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y~(x)) +
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
# specify the text (i.e., label) and the x- and y-coordinates for text placement
annotate(geom = "text", x = 250, y = 2, label = "R2 = ???, p = ???", color = "red") +
my.theme
if your data set is continually changing (say, as you run more participants), you must continually hand correct the annotations
this is cumbersome, and it’s easy to make mistakes
SOLUTION: supply the label argument with output directly from regression model
let’s see an example
# Recall, we had previously specified geom_smooth(method='lm', formula=y~(x)).
# The call below produces the same model, just outside of ggplot.
my.model <- summary(lm(wt ~ hp, data = df1))
print(my.model)
Call:
lm(formula = wt ~ hp, data = df1)
Residuals:
Min 1Q Median 3Q Max
-1.4176 -0.5312 -0.0204 0.4254 1.5645
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.83825 0.31652 5.81 2.4e-06 ***
hp 0.00940 0.00196 4.80 4.1e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.748 on 30 degrees of freedom
Multiple R-squared: 0.434, Adjusted R-squared: 0.415
F-statistic: 23 on 1 and 30 DF, p-value: 4.15e-05
# extract the R-squared value for the weight~horsepower correlation
my.model.r2 = my.model$r.squared
print(my.model.r2)[1] 0.4339# combine R-squared value into a descriptive string
my.exp <- paste("R2 =", my.model.r2, sep = " ")
print(my.exp)[1] "R2 = 0.433948779081183"ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y~(x)) +
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000)", limits = c(1,6)) +
# We no longer specify the annotation text locally.
# The text comes directly from the regression model.
annotate(geom = "text", x = 250, y = 2, label = my.exp, color = "red")+
my.theme
lm_eqn = function(df1){
m = lm(wt ~ hp, df1);
eq <- substitute(italic(y) == a + b %.% italic(x)*","
~~italic(r)^2~"="~r2~~", p < "~~p,
list(a = format(coef(m)[1], digits = 2),
b = format(coef(m)[2], digits = 2),
r2 = format(summary(m)$r.squared, digits = 3),
p = format(coef(summary(m))[8], digits = 3, scientific=FALSE)
))
as.character(as.expression(eq));
}Credit goes to Ramnath Vaidyanathan for this function. Go here for the original post on StackOverflow.
ggplot(data = df1, aes(x = hp, y = wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y~(x)) +
scale_x_continuous("gross horsepower") +
scale_y_continuous("weight (lb/1000", limits = c(1,6)) +
annotate(geom = "text", x = 250, y = 2, label = lm_eqn(df1), parse = T, color = "red") +
my.theme
Grouping and Statistically Transforming Data
df2 <- subset(diamonds, carat > 0.5)
head(df2) carat cut color clarity depth table price x y z
91 0.70 Ideal E SI1 62.5 57 2757 5.70 5.72 3.57
92 0.86 Fair E SI2 55.1 69 2757 6.45 6.33 3.52
93 0.70 Ideal G VS2 61.6 56 2757 5.70 5.67 3.50
94 0.71 Very Good E VS2 62.4 57 2759 5.68 5.73 3.56
95 0.78 Very Good G SI2 63.8 56 2759 5.81 5.85 3.72
96 0.70 Good E VS2 57.5 58 2759 5.85 5.90 3.38nrow(df2)[1] 35008ggplot(data = df2, aes(x = carat, y = price)) +
geom_point() +
geom_smooth(method = "lm") +
scale_x_continuous("carat") +
scale_y_continuous("price")+
my.theme
ggplot(data = df2, aes(x = log10(carat), y = log10(price))) +
geom_point() +
geom_smooth(method = "lm") +
scale_x_continuous("log10(carat)") +
scale_y_continuous("log10(price)") +
my.theme
ggplot(data = df2, aes(x = carat, y = price)) +
geom_point() +
geom_smooth(method = "lm") +
coord_trans(x = "log10", y = "log10") +
scale_x_continuous("log10(carat)") +
scale_y_continuous("log10(price)") +
my.theme
ggplot(data=df2, aes(x=carat, y=price, colour=cut, fill=cut)) +
geom_point() +
geom_smooth(method = "lm") +
coord_trans(x = "log10", y = "log10") +
scale_x_continuous("log10(carat)") +
scale_y_continuous("log10(price)") +
my.theme
ggplot(data=df2, aes(x=carat, y=price, colour=cut, fill=cut)) +
geom_point(alpha = .05) + # number btw 0 (transparent) and 1 (opaque)
geom_smooth(method = "lm") +
coord_trans(x = "log10", y = "log10") +
scale_x_continuous("log10(carat)") +
scale_y_continuous("log10(price)") +
my.theme
ggplot(data=df2, aes(x=carat, y=price, colour=cut, fill=cut)) +
geom_point(alpha = .05) +
geom_smooth(method = "lm") +
coord_trans(x = "log10", y = "log10") +
scale_x_continuous("log10(carat)") +
scale_y_continuous("log10(price)") +
guides(fill = guide_legend(reverse = TRUE), colour = guide_legend(reverse = TRUE)) +
my.theme
# plot number of tokens (counts) that fall within a specific bin
ggplot(data=df2, aes(x=price, fill=cut, colour=cut)) +
geom_histogram(alpha=.8) +
guides(fill = guide_legend(reverse = TRUE), colour = guide_legend(reverse = TRUE)) +
my.theme
# plot the probability that a variable will take on a given value
ggplot(data=df2, aes(x=price, fill=cut, colour=cut)) +
geom_density(alpha=.5) +
guides(fill = guide_legend(reverse = TRUE), colour = guide_legend(reverse = TRUE)) +
my.theme
# the trick here is to change the y-axis for the histrogram
# from counts to probability density
ggplot(data=df2, aes(x=price)) +
geom_histogram(aes(y=..density..), alpha=.5) +
geom_density(size=1.25) +
my.theme
ggplot(data=df2, aes(x=price)) +
geom_histogram(aes(y=..density..), alpha=.5) +
geom_density(size=1.25, adjust=1.9) +
my.theme
ggplot(data=df2, aes(x=price, fill=..count..)) +
geom_histogram() +
scale_fill_continuous(low="violet", high="violetred4") +
my.theme
# define subset for easy reference
df2.sub <- subset(df2, cut %in% c("Ideal","Good") & price < 8000)
ggplot(data=df2.sub, aes(x=price, colour=cut)) +
geom_histogram(aes(y=..density.., fill=cut), alpha=.3) +
geom_density(size=1.5) +
facet_grid(cut ~ .) +
my.theme
# calculate means of each group
df2.sub.means <- ddply(df2.sub, .(cut),
summarise, mean.price=mean(price))Error: could not find function "ddply"
print(df2.sub.means)Error: object 'df2.sub.means' not found
ggplot(data=df2.sub, aes(x=price, colour=cut)) +
geom_histogram(aes(y=..density.., fill=cut), alpha=.3) +
geom_density(size=1.5) +
geom_vline(data=df2.sub.means, aes(xintercept=mean.price),
linetype="dashed", size=1, colour="red") + # add means as dashed lines
facet_grid(cut ~ .) + # plot subgroups as separate "facets"
my.themeError: object 'df2.sub.means' not foundSome Cosmetic Details
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
my.theme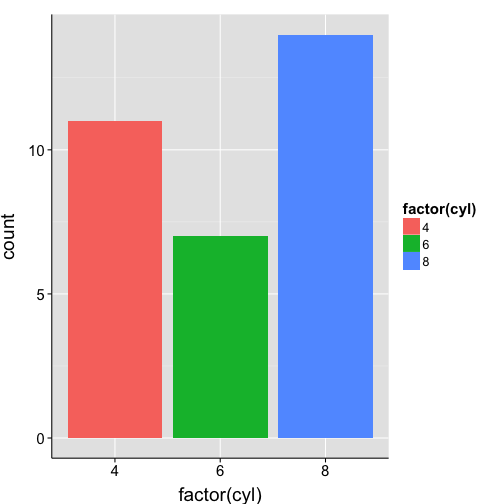
ggplot(mpg, aes(x = trans, fill = trans)) +
geom_bar() +
my.theme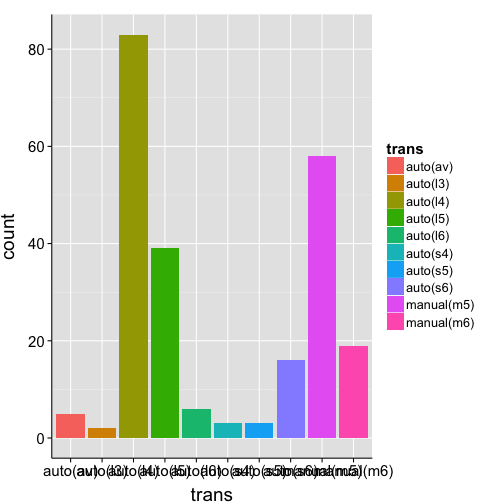
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_manual(values=c("red", "green", "blue")) +
my.theme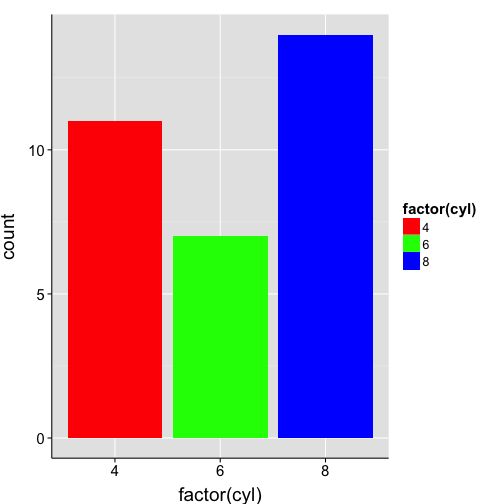
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_manual(values=c("#980000", "#009800", "#000098")) +
my.theme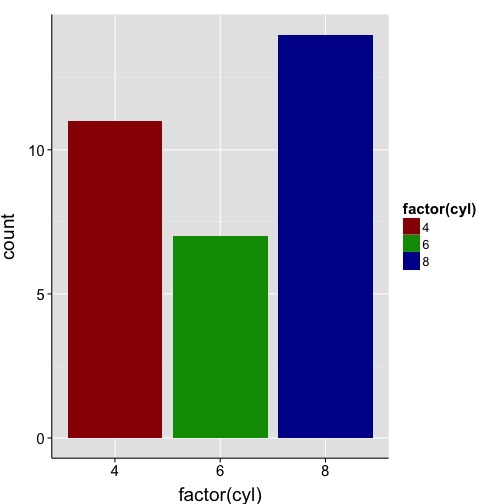
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_brewer() +
my.theme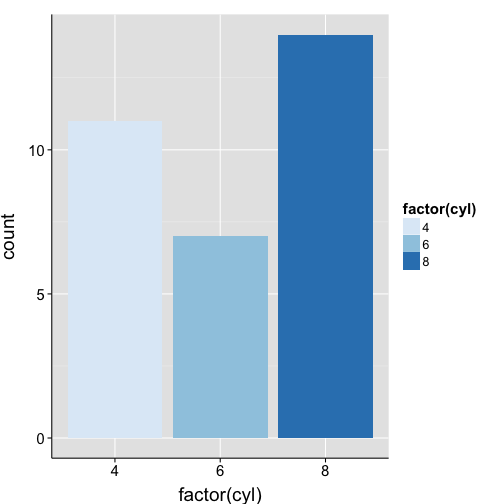

brewer
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_brewer(palette="YlOrRd")+
my.theme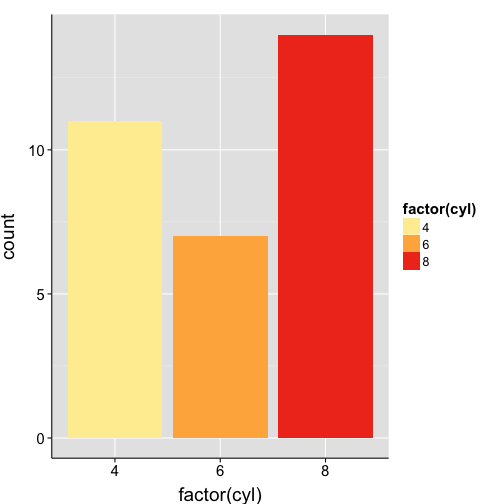
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_grey() +
my.theme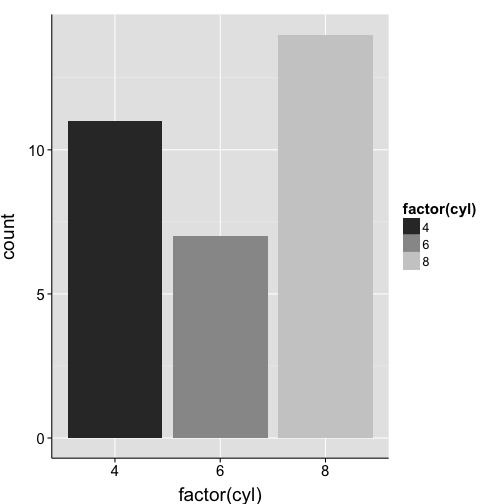
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_grey("Cylinders") +
my.theme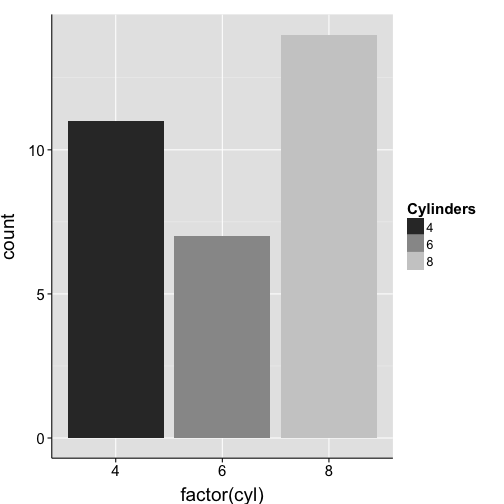
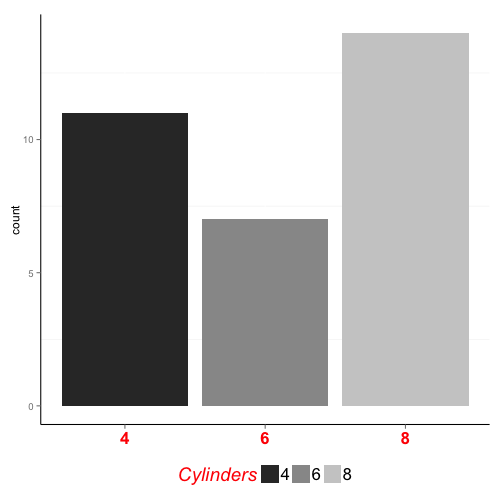
ggplot(mtcars, aes(x = factor(cyl), fill = factor(cyl))) +
geom_bar() +
scale_fill_grey("Cylinders") +
theme(legend.position="bottom", # move legend below x-axis
legend.title = element_text(colour="red", face="italic", size=19),
legend.text = element_text(size=17),
axis.text.x = element_text(colour="red", face="bold", size=17),
axis.title.x = element_blank(), # remove x-axis label
panel.background = element_blank(), # remove panel grid
axis.line = element_line() # add 'l-shaped' axis line
) 
Linguistic Data
Part 1: Formant Values and Vowel Plots
# acoustic measurements of 8 reps of 10 vowels from 72 speakers (adults and
# children)
pb <- read.delim("http://ling.upenn.edu/courses/cogs501/pb.Table1", header = FALSE)
# Give meaningful column names
colnames(pb) <- c("Age", "Sex", "Speaker", "Vowel", "V1", "F0.hz", "F1.hz", "F2.hz",
"F3.hz")
# Relabel the age data
levels(pb$Age) <- c("Child", "Adult", "Adult")
pb$Age <- relevel(pb$Age, "Adult")
# Clean up speaker labeling
pb$Speaker <- as.factor(paste("S", pb$Speaker, sep = ""))
head(pb) Age Sex Speaker Vowel V1 F0.hz F1.hz F2.hz F3.hz
1 Adult m S1 iy i 160 240 2280 2850
2 Adult m S1 iy i 186 280 2400 2790
3 Adult m S1 ih \\ic 203 390 2030 2640
4 Adult m S1 ih \\ic 192 310 1980 2550
5 Adult m S1 eh \\ep 161 490 1870 2420
6 Adult m S1 eh \\ep 155 570 1700 2600# note we can select a subset from within the call to ggplot
ggplot(data=subset(pb, Speaker=="S2"), aes(x=F2.hz, y=F1.hz, label=Vowel)) +
geom_point() +
geom_text() + # geom_text() labels points. Label must be specified as an aesthetic
scale_y_reverse() + # reverse the axes
scale_x_reverse() +
my.theme
# note we can select a subset from within the call to ggplot
ggplot(data=subset(pb, Speaker=="S2"), aes(x=F2.hz, y=F1.hz, label=Vowel)) +
geom_point() +
geom_text(hjust=1.1, vjust=-.2, size=5.5) + # adjust placement and size of labels
scale_y_reverse() +
scale_x_reverse() +
my.theme
ggplot(data=subset(pb, Speaker %in% c("S1","S2","S3","S4")), aes(x=F2.hz, y=F1.hz, label=Vowel)) +
geom_text(hjust=1, vjust=-.2) +
scale_y_reverse() +
scale_x_reverse() +
facet_wrap(~ Speaker) + # this line provides functionality similar to the lattice package
my.theme
ggplot(data=pb, aes(x=F2.hz, y=F1.hz, label=Vowel, colour=Vowel)) +
geom_text(hjust=1, vjust=-.2) +
scale_y_reverse() +
scale_x_reverse() +
facet_wrap(~ Sex + Age) + # specify multiple variables for faceting
my.theme
Note: some male-adult values are being cut off
ggplot(data=pb, aes(x=F2.hz, y=F1.hz, label=Vowel, colour=Vowel)) +
geom_text(hjust=1, vjust=-.2) +
scale_y_reverse() +
scale_x_reverse() +
coord_cartesian(ylim=c(0, 1500)) +
facet_wrap(~ Sex + Age) +
my.theme
There are many ways to adjust axis limits, but when axes are reversed, it’s best to use coord_cartesian()
ggplot(data=subset(pb, Sex=="f"), aes(x=F2.hz, y=F1.hz, label=Vowel, colour=Age)) +
geom_text() +
scale_y_reverse() +
scale_x_reverse() +
my.theme
… we could plot shaded regions showing the total area of the child’s formant space vs. the adult’s space?
as it turns out, geom_polygon can do just that
first, we just need to find the envelope (i.e., convex hull) for adults and children
require(plyr)
# chull() is a function for finding the 'envelope' of a set of data points.
# Here we use this function to find the extreme points for a 2-dimensional
# space (F1-F2)
find_hull <- function(df) {
df[chull(df$F2.hz, df$F1.hz), ]
}
# apply this function to find the envelope for females by Age (adult vs.
# children)
hulls <- ddply(subset(pb, Sex == "f"), .(Age), find_hull)
print(hulls) Age Sex Speaker Vowel V1 F0.hz F1.hz F2.hz F3.hz
1 Adult f S54 iy i 220 220 2850 3800
2 Adult f S37 ah \\vt 213 280 850 2500
3 Adult f S61 ah \\vt 260 290 670 2380
4 Adult f S61 ah \\vt 275 330 630 2460
5 Adult f S59 uh \\hs 246 420 590 3100
6 Adult f S58 ao \\ct 250 950 1130 3160
7 Adult f S53 ao \\ct 267 987 1172 3180
8 Adult f S40 ao \\ct 180 1040 1300 3000
9 Adult f S48 ae \\ae 222 1110 2160 2700
10 Adult f S40 iy i 200 300 3100 3400
11 Child f S63 iy i 290 320 3500 4260
12 Child f S68 ah \\vt 260 310 730 3500
13 Child f S68 ah \\vt 274 360 660 3050
14 Child f S62 ah \\vt 200 400 650 3800
15 Child f S66 uh \\hs 285 770 940 3750
16 Child f S76 ao \\ct 350 1190 1470 3150
17 Child f S62 ao \\ct 200 1300 1800 3450
18 Child f S67 ae \\ae 214 1240 2700 3640
19 Child f S67 iy i 227 590 3610 4220ggplot(data=subset(pb, Sex=="f"), aes(x=F2.hz, y=F1.hz, label=Vowel, colour=Age, fill=Age)) +
geom_text() +
geom_polygon(data=hulls, alpha=.4) +
scale_y_reverse() +
scale_x_reverse() +
my.theme
Linguistic Data
Part 2: Response Times, Barplots, and Error Bars
standard error of the mean equals std. deviation (\(\sigma\)) divided by the square root of the sample size (N)
\[ SE_{\bar{x}} = \sigma / \sqrt{N} \]
which is equivalent to square root of variance (\(s^2\)) divided by square root of sample size (N)
\[ \sqrt{s^2 / N} \]
std.error <- function(x, na.rm = T) {
sqrt(var(x, na.rm = na.rm)/length(x[complete.cases(x)]))
}# read in fake data
rt.dat <- read.delim("datasets/fakedata.txt", header = T)
head(rt.dat) Subject Item IV1 IV2 Response Freq RT
1 S1 t_01 A 2 1 0.24 1100
2 S1 t_02 A 2 1 5.22 983
3 S1 t_03 A 2 1 5.10 1108
4 S1 t_04 A 2 1 5.77 984
5 S1 t_05 A 2 1 2.37 1340
6 S1 t_06 A 2 1 5.80 1192# How many data points per subjet per condition
table(rt.dat$IV1, rt.dat$IV2)/length(unique(rt.dat$Subject))
2 3 4
A 8 8 8
B 8 8 8require(plyr)
# use ddply() to calculate condition means for each subject, averaging over items
mean.bySubj <- ddply(rt.dat, .(Subject, IV1, IV2), summarise,
"mean.RT" = mean(RT))
head(mean.bySubj, 18) Subject IV1 IV2 mean.RT
1 S1 A 2 1150
2 S1 A 3 1309
3 S1 A 4 1453
4 S1 B 2 1067
5 S1 B 3 1479
6 S1 B 4 1694
7 S10 A 2 1245
8 S10 A 3 1318
9 S10 A 4 1636
10 S10 B 2 1136
11 S10 B 3 1467
12 S10 B 4 1914
13 S11 A 2 1234
14 S11 A 3 1418
15 S11 A 4 1679
16 S11 B 2 1102
17 S11 B 3 1435
18 S11 B 4 1894grand.means <- ddply(mean.bySubj, .(IV1, IV2), summarise,
"grand.RT" = mean(mean.RT),
"se" = std.error(mean.RT))
print(grand.means) IV1 IV2 grand.RT se
1 A 2 713.9 76.75
2 A 3 885.9 76.33
3 A 4 1152.3 77.98
4 B 2 721.9 74.06
5 B 3 1019.1 81.86
6 B 4 1445.9 77.44ggplot(data = grand.means, aes(x = factor(IV2), y = grand.RT, fill = IV1)) +
geom_bar(position = position_dodge(.9)) + # position bars side-by-side (cf. stacked)
geom_errorbar(position = position_dodge(.9), width = .25,
aes(ymin = grand.RT-se, ymax = grand.RT+se)) + # ymax and ymin are group mean +/- std. error
my.theme
ggplot(data = grand.means, aes(x = factor(IV2), y=grand.RT, group=IV1, colour=IV1)) +
geom_point() + # use points and lines instead of geom_bar()
geom_line() +
# note: we no longer need to specify position_dodge for the errorbars
geom_errorbar(width = .15, aes(ymin = grand.RT-se, ymax = grand.RT+se)) +
my.theme
ggplot(data = grand.means, aes(x = factor(IV2), y=grand.RT, group=IV1, colour=IV1)) +
geom_point() +
geom_line(linetype="dashed") +
geom_errorbar(width = .15, aes(ymin = grand.RT-se, ymax = grand.RT+se)) +
my.theme
# immediately calculate condition means and error from raw data,
# without first averaging over subjs
means.wrong <- ddply(rt.dat, .(IV1, IV2), summarise, "mean.RT" = mean(RT))
error.wrong <- ddply(rt.dat, .(IV1,IV2), summarise, "se" = std.error(RT))
# merge means and error into one frame
wrong.grand <- merge(means.wrong, error.wrong, by=c("IV1", "IV2"), all=T)
print(wrong.grand) IV1 IV2 mean.RT se
1 A 2 713.9 28.56
2 A 3 885.9 29.06
3 A 4 1152.3 30.37
4 B 2 721.9 27.55
5 B 3 1019.1 31.51
6 B 4 1445.9 32.02
The error bars became substantially smaller. Why?
Recall that when calculating standard error of the mean correctly, the denominator is the sample size of the data set (e.g., number of participants)
Hence, we want to calculate error using a data frame containing one data point per subject per condition (i.e., each subject’s mean performance in that condition)
If we calculate standard error over the whole data set (as in the second example above), instead of over subject means, then the denominator is huge! Which leads to a tiny error measurement.
Linguistic Data
Part 3: Binomial DVs and Proportions
## lexdec is a canned lexical decision data set from Harold Baayen and Jen Hay
## available in the package languageR
# install.packages('languageR')
require(languageR)
# numeric coding of of whether response was correct or incorrect
lexdec$Correct.num <- ifelse(lexdec$Correct == "correct", 1, 0)
# simplify column names
colnames(lexdec)[c(5, 10)] <- c("NativeLang", "Freq")
# select specific subset for illustrative purposes
subjs <- c("A1", "A2", "C", "K", "R3", "S", "T1", "J", "M2", "T2", "V", "Z")
lex.sub <- subset(lexdec, Subject %in% subjs)
str(lex.sub)'data.frame': 948 obs. of 29 variables:
$ Subject : Factor w/ 21 levels "A1","A2","A3",..: 1 1 1 1 1 1 1 1 1 1 ...
$ RT : num 6.34 6.31 6.35 6.19 6.03 ...
$ Trial : int 23 27 29 30 32 33 34 38 41 42 ...
$ Sex : Factor w/ 2 levels "F","M": 1 1 1 1 1 1 1 1 1 1 ...
$ NativeLang : Factor w/ 2 levels "English","Other": 1 1 1 1 1 1 1 1 1 1 ...
$ Correct : Factor w/ 2 levels "correct","incorrect": 1 1 1 1 1 1 1 1 1 1 ...
$ PrevType : Factor w/ 2 levels "nonword","word": 2 1 1 2 1 2 2 1 1 2 ...
$ PrevCorrect : Factor w/ 2 levels "correct","incorrect": 1 1 1 1 1 1 1 1 1 1 ...
$ Word : Factor w/ 79 levels "almond","ant",..: 55 47 20 58 25 12 71 69 62 1 ...
$ Freq : num 4.86 4.61 5 4.73 7.67 ...
$ FamilySize : num 1.386 1.099 0.693 0 3.135 ...
$ SynsetCount : num 0.693 1.946 1.609 1.099 2.079 ...
$ Length : int 3 4 6 4 3 10 10 8 6 6 ...
$ Class : Factor w/ 2 levels "animal","plant": 1 1 2 2 1 2 2 1 2 2 ...
$ FreqSingular: int 54 69 83 44 1233 26 50 63 11 24 ...
$ FreqPlural : int 74 30 49 68 828 31 65 47 9 42 ...
$ DerivEntropy: num 0.791 0.697 0.475 0 1.213 ...
$ Complex : Factor w/ 2 levels "complex","simplex": 2 2 2 2 2 1 1 2 2 2 ...
$ rInfl : num -0.31 0.815 0.519 -0.427 0.398 ...
$ meanRT : num 6.36 6.42 6.34 6.34 6.3 ...
$ SubjFreq : num 3.12 2.4 3.88 4.52 6.04 3.28 5.04 2.8 3.12 3.72 ...
$ meanSize : num 3.48 3 1.63 1.99 4.64 ...
$ meanWeight : num 3.18 2.61 1.21 1.61 4.52 ...
$ BNCw : num 12.06 5.74 5.72 2.05 74.84 ...
$ BNCc : num 0 4.06 3.25 1.46 50.86 ...
$ BNCd : num 6.18 2.85 12.59 7.36 241.56 ...
$ BNCcRatio : num 0 0.708 0.568 0.713 0.68 ...
$ BNCdRatio : num 0.512 0.497 2.202 3.591 3.228 ...
$ Correct.num : num 1 1 1 1 1 1 1 1 1 1 ...ggplot(data = lex.sub, aes(x=Freq, y=Correct.num, colour=NativeLang, fill=NativeLang)) +
geom_point() +
# fit data with a logistic regression line
geom_smooth(method="glm", family="binomial", formula=y~(x)) +
scale_y_continuous("Proportion correct") +
scale_x_continuous("Word Frequency") +
my.theme
ggplot(data = lex.sub, aes(x=Freq, y=Correct.num, colour=NativeLang, fill=NativeLang)) +
geom_point() +
geom_smooth() +
scale_y_continuous("Proportion correct") +
scale_x_continuous("Word Frequency") +
my.theme
# calculate proportion correct for each subject as a function of whether the
# previous type was a word or nonword
props.bySubj <- ddply(lex.sub, .(Subject, NativeLang, PrevType), function(x) {
sum(x$Correct.num)/nrow(x)
})
# change name of the proportion columns
names(props.bySubj)[names(props.bySubj) == "V1"] <- "Prop"
print(props.bySubj) Subject NativeLang PrevType Prop
1 A1 English nonword 0.9767
2 A1 English word 1.0000
3 A2 English nonword 0.9487
4 A2 English word 0.9750
5 C English nonword 1.0000
6 C English word 0.9722
7 J Other nonword 0.8667
8 J Other word 0.8235
9 K English nonword 1.0000
10 K English word 0.8537
11 M2 Other nonword 0.9444
12 M2 Other word 0.9535
13 R3 English nonword 0.9750
14 R3 English word 0.9744
15 S English nonword 0.9750
16 S English word 1.0000
17 T1 English nonword 0.9487
18 T1 English word 0.9750
19 T2 Other nonword 0.9048
20 T2 Other word 0.9189
21 V Other nonword 0.9773
22 V Other word 0.9429
23 Z Other nonword 0.9111
24 Z Other word 0.9118ggplot(data = props.bySubj, aes(x = PrevType, y = Prop, colour = NativeLang)) +
stat_summary(fun.data = "mean_cl_boot", size = 1.4) +
my.theme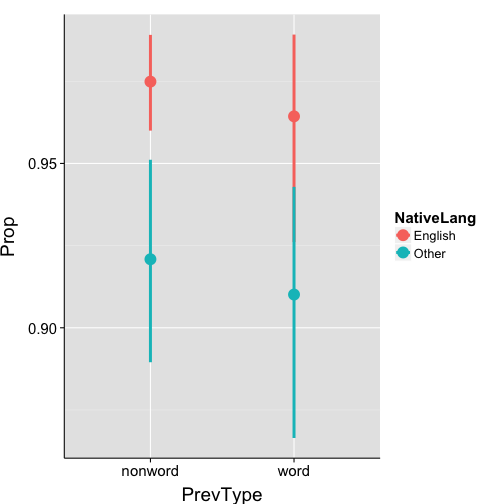
ggplot(data = props.bySubj, aes(x = PrevType, y = Prop, fill = NativeLang)) +
stat_summary(position=position_dodge(.9), fun.y=mean, geom="bar") +
stat_summary(position=position_dodge(.9), fun.data = "mean_cl_boot", geom="errorbar", width=.25, colour="black") +
my.theme
# calculate grand mean and standard error by native language and previous word
# type
grand.props <- ddply(props.bySubj, .(NativeLang, PrevType), summarise, mean.prop = mean(Prop),
se = std.error(Prop))
print(grand.props) NativeLang PrevType mean.prop se
1 English nonword 0.9749 0.00792
2 English word 0.9643 0.01901
3 Other nonword 0.9209 0.01875
4 Other word 0.9101 0.02295ggplot(data = grand.props, aes(x = PrevType, y = mean.prop, fill = NativeLang)) +
geom_bar(position = position_dodge(.9)) +
geom_errorbar(position = position_dodge(.9), width = .25,
aes(ymin = mean.prop-se, ymax = mean.prop+se)) +
my.theme
Linguistic Data
Part 4: Geospatial Plotting and Possibilities for Sociolinguistics and Dialectology
# install.packages("maps")
require(maps)
# collect longitude and latitude coordinates for US states
states <- map_data("state")
head(states) long lat group order region subregion
1 -87.46 30.39 1 1 alabama <NA>
2 -87.48 30.37 1 2 alabama <NA>
3 -87.53 30.37 1 3 alabama <NA>
4 -87.53 30.33 1 4 alabama <NA>
5 -87.57 30.33 1 5 alabama <NA>
6 -87.59 30.33 1 6 alabama <NA>ggplot(data=states, aes(map_id = region, group="group")) +
geom_map(map = states) +
expand_limits(x = states$long, y = states$lat)
ggplot(data=states, aes(map_id = region, group="group")) +
# colour affects border, fill affects state regions
geom_map(color="#757575", fill="lightgray", map = states) +
expand_limits(x = states$long, y = states$lat) +
coord_map() 
geo.dat <- read.delim("datasets/fakedata_geo.txt", header=T)
head(geo.dat) lat long region group
1 41.6 -80.9 ohio B
2 42.1 -83.9 ohio D
3 39.7 -84.2 ohio A
4 40.6 -82.4 ohio B
5 39.7 -82.3 ohio D
6 41.9 -82.5 ohio Astr(geo.dat)'data.frame': 323 obs. of 4 variables:
$ lat : num 41.6 42.1 39.7 40.6 39.7 41.9 41.4 38.6 42.3 39.5 ...
$ long : num -80.9 -83.9 -84.2 -82.4 -82.3 -82.5 -81.7 -84.1 -82.7 -82.2 ...
$ region: Factor w/ 10 levels "georgia","idaho",..: 6 6 6 6 6 6 6 6 6 6 ...
$ group : Factor w/ 5 levels "A","B","C","D",..: 2 4 1 2 4 1 1 1 1 2 ...ggplot(data=states, aes(map_id = region, group="group")) +
geom_map(color="#757575", fill="lightgray", map = states) +
expand_limits(x = states$long, y = states$lat) +
coord_map() +
geom_point(data = geo.dat, aes(x=long, y=lat, colour=group)) # HERE'S THE MAGIC! 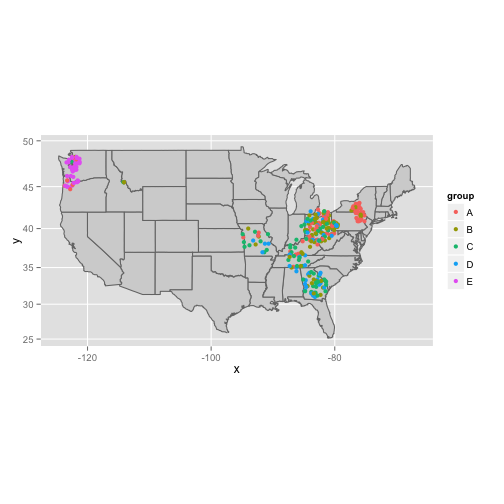
ggplot(data=states, aes(map_id = region, group="group")) +
geom_map(color="#757575", fill="lightgray", map = states) +
expand_limits(x = states$long, y = states$lat) +
coord_map() +
geom_point(data = geo.dat, aes(x=long, y=lat, colour=group)) +
theme(panel.background = element_blank(), # remove panel background
axis.ticks = element_blank(),
axis.text = element_blank(),
axis.title = element_blank())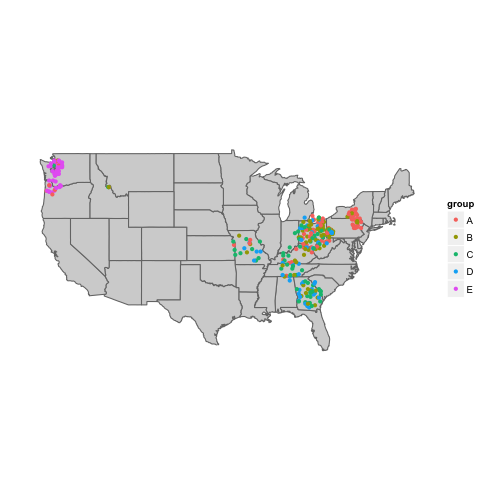
Thanks to: